Genome assemblies with HiFi data
NGI can generate high quality assemblies using IPA and hifiasm assemblers
Improved Phased Assembler (IPA) is the official PacBio software for HiFi genome assembly. IPA was designed to utilize the accuracy of PacBio HiFi reads to produce high-quality phased genome assemblies.
Hifiasm is a fast haplotype-resolved de novo assembler for PacBio HiFi reads. It emits partially phased assemblies of quality competitive with the best assemblers. Given parental short reads or Hi-C data, it produces arguably the best haplotype-resolved assemblies so far.
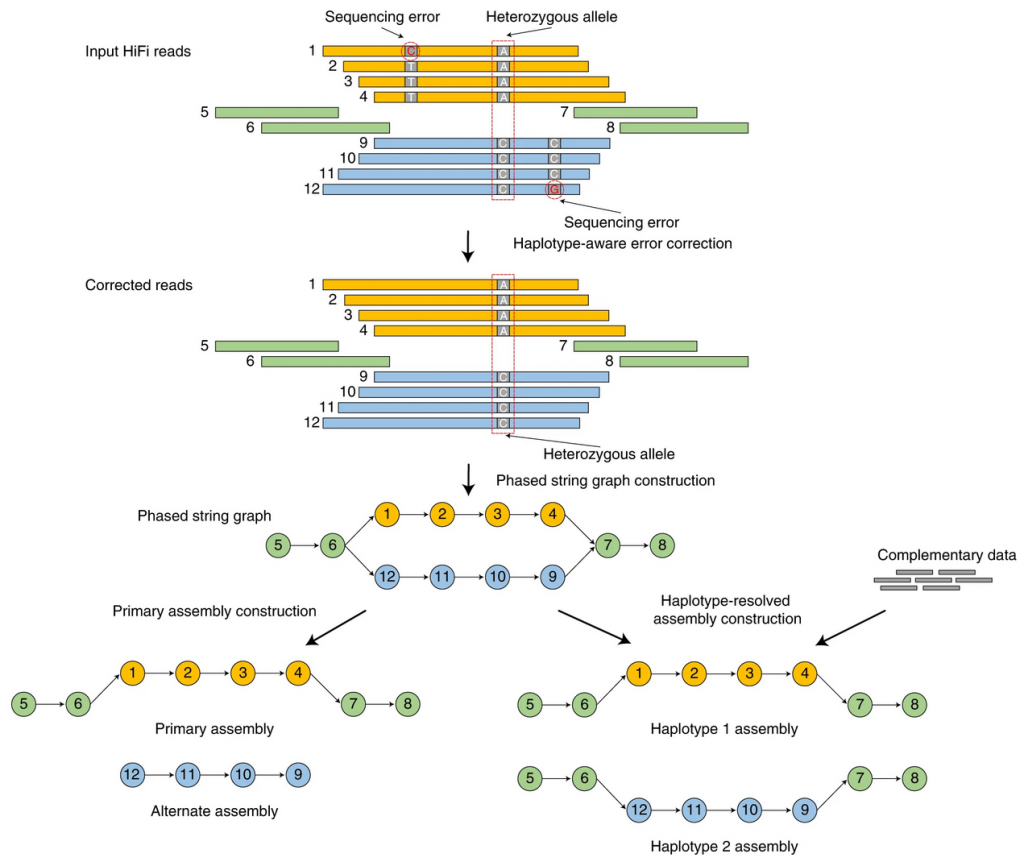
NGI provides de novo assemblies using both methods. We recommend to generate 15-20X of HiFi data per haplotype. Coverage requirements scale linearly by the number of unique haplotypes. For example, a highly heterozygous diploid may require double the recommended coverage. While a homozygous tetraploid may also require double coverage, (in a case where haplotypes are identical, but homeologs are not).
We highly recommend to complement HiFi assemblies with high-quality proximity ligation data generated using methods such as Omni-C. Combination of both data types allows generation of phased, chromosome level genome assemblies.
NGI can provide scaffolding service and help with initial manual curation. Please contact us for more information regarding this service.
Last Updated: 29th September 2022