Index guidelines for Illumina sequencing
Indexes, also known as barcodes, are used to bioinformatically identify which read corresponds to which sample after sequencing. These short index sequences are attached to each sample during the library preparation, serving as a unique identifier for that specific sample. Typically, dual indexes are used during library preparation, with one index on each side of the sequence of interest, referred to as i5 and i7 indexes. When choosing indexes they can either all be unique, Unique Dual Indexes (UDI), or have unique combinations of fewer different indexes, Combinatorial Dual Indexes (CDI).
For additional details and sequencing specification, please refer to the latest documentation on Illuminas website:
- Index Adapters Pooling Guide: https://emea.support.illumina.com/downloads/index-adapters-pooling-guide-1000000041074.html
- Indexed Sequencing on Illumina Systems: https://knowledge.illumina.com/library-preparation/general/library-preparation-general-reference_material-list/000002099
There are two different strategies used for Illumina sequencing. The first one is called the forward strand workflow and is used for sequencing on instruments such as the MiSeq system.
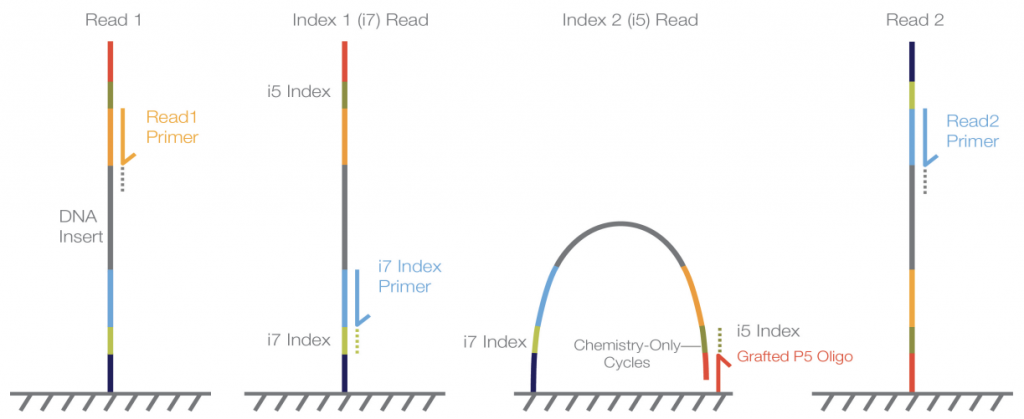
This workflow starts by sequencing of Read 1 after annealing of the Read1 primer, that reads the target template (insert) and washes the read product off before adding the i7 Index primer for sequencing of the i7 Index. The entire fragment then folds and is sequenced from the grafted P5 oligo into the i5 Index and synthesizes the reverse complement. Lastly, the Read 2 primer and insert are read.
The second strategy is the reverse complement workflow, which is used for sequencing on instruments such as NovaSeq X Plus, NovaSeq 6000 (v1.5), and NextSeq 2000.
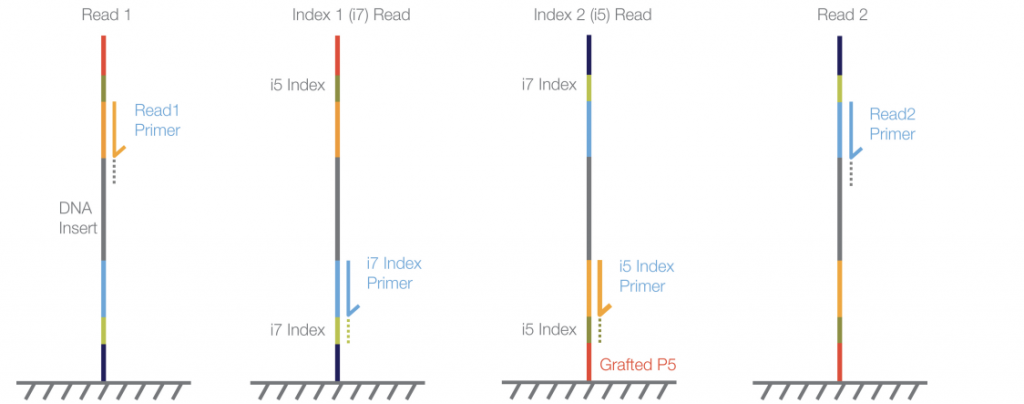
This method starts Read 1 at the read 1 primer, reads the inserts and washes the read product off before reading the i7 index primer and i7 index. The entire fragment then folds and is read from the grafted p5 oligo to synthesize the reverse complement, and off of this then reads the i5 index primer and i5 index. This read product is then washed off before reading the Read 2 primer and insert.
Of note, in the the reverse complement workflow, the i5 index is read after the index primer, whereas the forward strand method reads the i5 index after the grafted oligo. Therefore, the i5 index sequence needs to be specified in either the forward direction or the reverse complement direction depending on the workflow, which varies between the different sequencing machines.
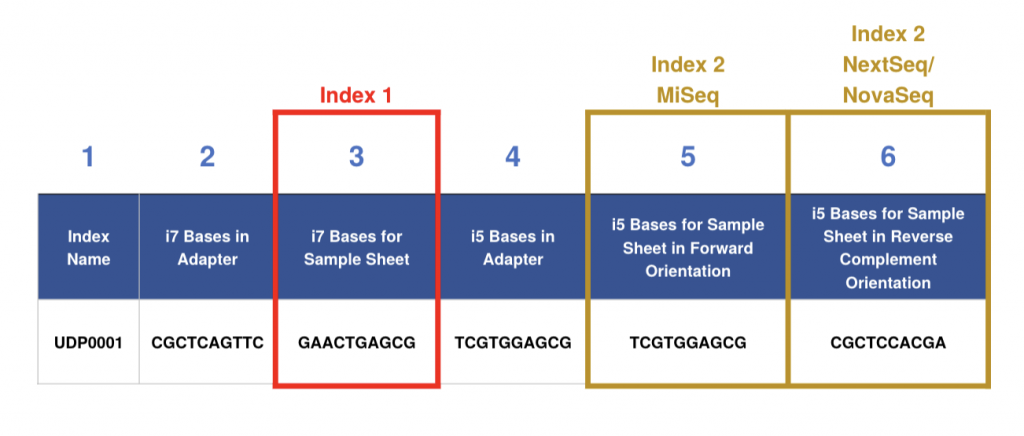
If you are using a commercial indexing kit to make your libraries, the kit documentation will most likely include the index name, the i7 bases in adapter and i5 bases in adapter, corresponding to columns 1, 2 and 4 in the table above.
For the submission sample sheet, we need to know the i7 bases for sample sheet (column 3) and either column 5 or 6 depending on what sequencing instrument is used (as described above).
The i5 sequence as seen on the index kit is needed for machines using the forward strand workflow, at NGI that is the MiSeq. The other instruments, NextSeq 2000 and NovaSeq 6000/X Plus, use the reverse complement workflow, and therefore need the reverse complement of the i5 index bases on the sample sheet.
Example: If your sequencing library is labelled with the index “UDP0001″ as in the table above, for MiSeq the reported index should follow the format “i7 Bases for Sample Sheet-i5 Bases for Sample Sheet in Forward Orientation” (GAACTGAGCG-TCGTGGAGCG). For NovaSeq or Nextseq, the reported index shall follow the format “i7 Bases for Sample Sheet-i5 Bases for Sample sheet in Reverse Complement Orientation” (GAACTGAGCG-CGCTCCACGA)
Other considerations when choosing indexes include the index diversity and having optimal color balance in each sequencing cycle. On Illumina’s 2-channel chemistry, which is used on the NextSeq and NovaSeq systems, there may be issues reading the nucleotides if there is too little of one color’s signal, or if there are several cycles with no signal. This could be an issue if you use combinatorial indexes, and one or more cycles happen to have a low diversity due to specific combinations of repeated indexes. Index diversity can also be an issue if you are sequencing just a few libraries together.
Here is further information about this from Illumina.
If preparing sequencing libraries on your own, always check the index pooling recommendations from the company providing the library preparation kit, and Illuminas Index Adapters Pooling Guide: https://emea.support.illumina.com/downloads/index-adapters-pooling-guide-1000000041074.html
Last Updated: 16th April 2024