Transcriptomics
Methods to do bulk sequencing of protein coding, non-coding or small RNA, with short or long reads. Select an application to learn more about its advantages and sample requirements. We also offer different options for low input samples or degraded RNA within each application.
NGI offers several types of transcriptome sequencing approaches. Are you interested in analysing protein coding or non-coding RNAs, or small RNAs such as miRNAs? Get in touch with us to discuss the best approach for your project!
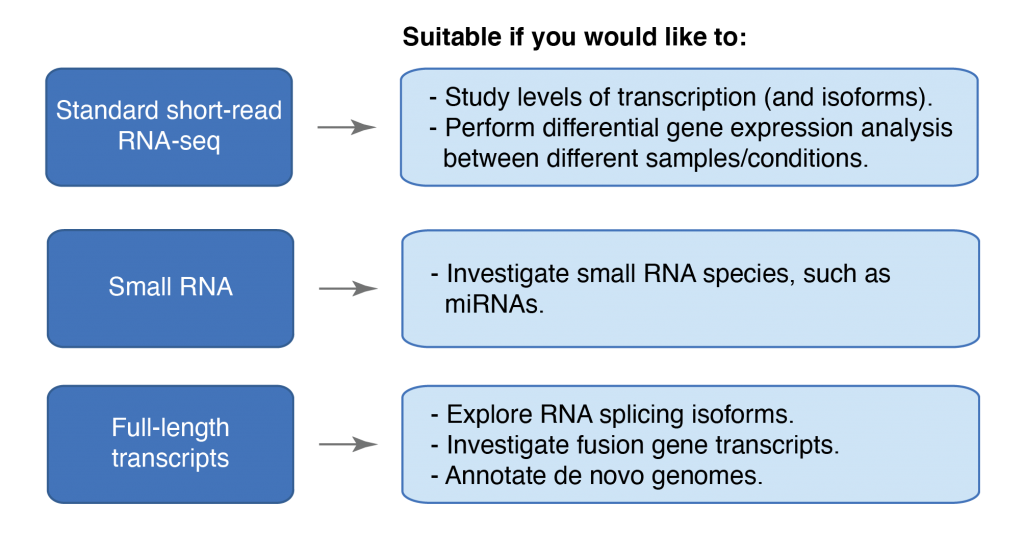
Please consult the flowchart for an overview of the applications that might best suit your project. You can also read more about each application further down in the applications section.
For projects focused on transcriptome annotation, please refer to our de novo page.
All new projects should first be discussed with us prior to placing an order through our online order portal. Please contact us here.
Library complexity
The term “coverage” is often used when sequencing DNA samples, referring to the average number of reads that align to, or "cover", known reference bases. When sequencing RNA samples, we are instead referring to “number of reads”, as transcripts are present at different levels.
For high quality RNA sequencing data, we ask for high-quality RNA samples in adequate amounts. Degraded or low amounts of starting material can yield low complexity libraries, meaning that the number of unique fragments present in a given library will be very low. This could complicate downstream analyses. It is therefore of utmost importance that our sample requirement guidelines are followed, and high quality RNA material is submitted to us.
General sample requirements
Each application has its own requirements regarding concentration and volume, as different starting amounts are needed. We strongly encourage our users to check sample amounts and quality before submitting them to us. Making sure you have good quality samples from the start will improve the success rate and facilitate handling of your project. Note that the quality control performed at NGI is mainly for confirmation purposes.
Concentration measurements are most accurately performed using fluorometric measurements (Qubit, Quant-it). For quality scoring we recommend a capillary electrophoresis method (Fragment Analyzer, Bioanalyzer, TapeStation) that can measure RNA integrity by providing a RNA integrity Number (RIN) value. RIN values range from 1–10, with lower values indicating poor quality and higher values indicating good quality RNA (RIN > 8). Please note that RIN values can be a bit tricky to obtain on certain types of RNA, such as insect RNA. Read more about RIN scores here and here. In cases where your project involves FFPE samples, we also recommend using a capillary electrophoresis method to determine the DV200 value.
The RNA should be free from any contaminating DNA to minimize the contribution of sequence reads derived from residual genomic DNA in the sample. NGI does not perform DNase treatment prior to library preparation. In particular for library preparation based on ribosomal depletion, failure to treat RNA samples with DNase or inefficient DNase treatment can result in a significant fraction of intergenic reads in the sequence data.
Primary data analysis
NGI supports primary RNA-seq analysis for Illumina (species listed in iGenomes) and PacBio applications.
Price examples
Price examples for RNA projects can be found here, select either the “Illumina sequencing” or the “Long read sequencing” tabs to learn more.
Standard short-read RNA-seq
Investigate transcription levels and isoforms, e.g. for studies of differential expression.Small RNA
Assess the abundance and identity of different small-RNA such as miRNA, directly from total RNA.Full-length transcripts
Full-length sequencing of RNA transcripts can be used to investigate splicing isoforms and fusion-gene transcripts, as well as annotation of de novo genomes.Illumina TruSeq Stranded mRNA
RNA sequencing of mRNAs selected through poly-A enrichment.
mRNA illumina RNA-Seq library preparation transcriptomics RNA truseqIllumina TruSeq Stranded RNA without selection or depletion
RNA sequencing of either all RNAs in a sample, or of a RNA sample depleted of for example rRNA by the user.
illumina rnaseq RNA-Seq library preparation RNA truseqIllumina Stranded total RNA Prep (Ribo-Zero Plus)
Total RNA sequencing based on reduced rRNA content and other type of highly abundant RNAs in both prokaryotic and eukaryotic samples.
library preparation transcriptomics RNA mRNA illumina RNA-Seq10X Genomics 3’ or 5’ gene expression (GEM-X or Next-GEM)
Profiling of gene expression levels at single-cell resolution.
transcriptomics single cell RNA mRNA illumina RNA-Seq chromium 10x Genomics library preparation10X Genomics Multiome (ATAC + gene expression)
Profiling of 3´gene expression and chromatin accessibility in the same cell.
ATAC-seq RNA-Seq ATAC chromium library preparation 10x Genomics transcriptomics single cell RNA mRNA illuminaFull-length transcript sequencing of 10X libraries with ONT sequencing
Combining short & long reads at the single cell level.
illumina ont chromium 10x Genomics full transcript library preparation single cell long-readIon Torrent sequencing
Ion Torrent semiconductor sequencing technology is as simple as it is fast.
amplicon ampliseq ion torrent s5Nanopore cDNA sequencing
Nanopore cDNA sequencing is able to sequence entire transcripts in one go, ideal for detecting isoforms and fusions events.
assembly long-read nanoporeNanopore Direct RNA sequencing
Nanopore direct RNA sequencing is able to sequence entire transcripts from native RNA, opening up opportunities to detect RNA modifications.
assembly long-read nanoporePacBio SMRT sequencing
PacBio SMRT sequencing generates reads tens of kilobases in length enabling high quality genome assembly, structural variant analysis, amplicon resequencing, full-length transcript isoform sequencing, full-length 16S rRNA sequencing and amplification free epigenetic characterization.
de novo iso seq sv revio smrt assembly pacbio methylation amplicon sequel hifi clrQIAseq miRNA
miRNA libraries from very low input total RNA samples & degraded total RNA.
library preparation epigenetics RNA illuminaTaKaRa SMARTer pico RNA kit
The Takara SMARTer Stranded Total RNA-Seq Kit v3 - Pico Input Mammalian kit is specifically designed for very low input total RNA samples. It also works with degraded total RNA.
library preparation transcriptomics RNA illumina rnaseq RNA-Seq depletionPacBio Iso-Seq Analysis
With long and accurate HiFi reads, you can characterize the full diversity of the transcriptome
pacbio iso-seq hifiPromethION secondary analysis
Additional compute intensive nanopore raw data processing services provided by NGI
methylation base modifications basecalling fast5Illumina QC analysis
Basic quality-control monitoring of Illumina FastQ sequence data.
fastqc fastq screen checkqc QCRNA-fusion analysis
Runs with illumina RNA-Seq data. Aligns to the reference genome, gives QC metrics, predicted gene fusions and finishes with graphically visualised reports.
fusion rnafusionRNA-seq analysis
Runs with illumina total RNA-sequencing data. Aligns to the reference genome, gives QC metrics and finishes with gene count matrices.
transcriptomics rnaseq RNA-SeqSmall-RNA analysis
Runs with illumina small RNA-sequencing data. Aligns to the reference genome, gives QC metrics and finishes with gene count matrices.
small RNA smrna-seq smrnaseq