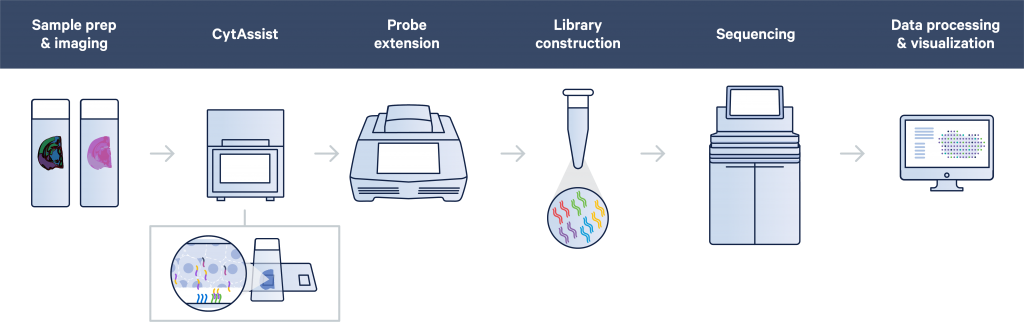
10X Genomics Visium CytAssist for Fresh Frozen and Fixed Frozen samples
We now offer spatially resolved transcriptomics through the 10X Genomics Visium CytAssist, which combines histology with probe-based transcriptomics in a spatial context.
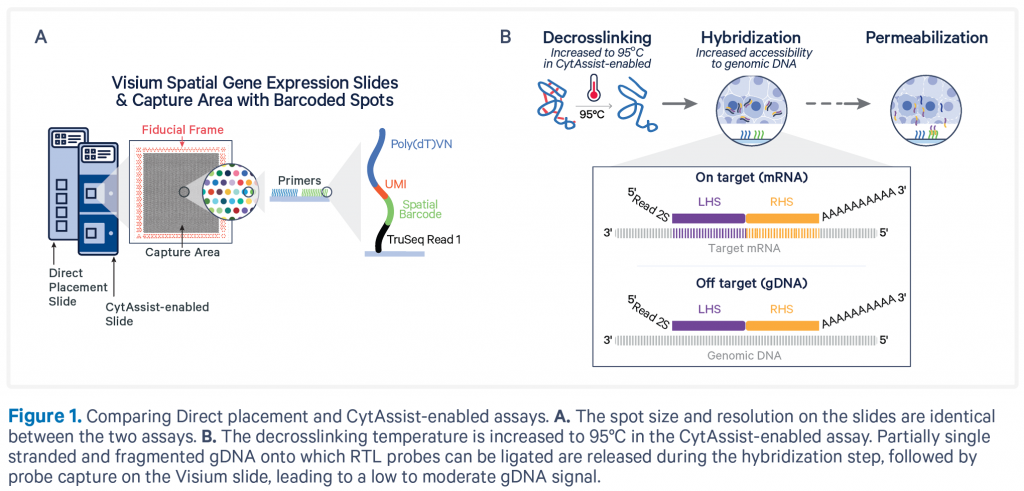
Coupling gene expression studies with tissue morphology is critical to our understanding of the biology and progression of disease. Similar to the FFPE – CytAssist protocol, the CytAssist protocol on Fresh Frozen (FF) or Fixed Frozen (FxF) tissues also uses the RNA-templated ligation (RTL) probe sets from 10X Genomics.
We offer these protocols in combination with the CytAssist instrument, which gives you the freedom to section your area of interest in your FF/FxF tissue samples in your own lab, place them on slides, and send those to us for processing.
Please contact us before planning your experiments for a discussion.
IMPORTANT! Do not section your samples without coordinating with us first. Libraries should be prepared within 2 months after sectioning so we need to ensure that we allocate time to perform this within this period.
Sample requirements
- The CytAssist protocol is probe-based and is so-far only available for human and mouse tissue samples.
- We require you to check that the morphology of the sections is good enough for cell type annotations. This can be assessed by H&E staining of sections consecutive to the ones you submit to NGI. Please send us the images that you obtain, highlighting your region of interest, prior to sample delivery (see image below). We will use this as a reference when selecting the areas from where the RNA should be transferred.
- Please extract RNA from the tissues that you use for sectioning. Estimate the RIN on a BioAnalyzer or similar; RIN should be ≥ 4. While low RIN scores do not necessarily result in poor data, high scores are more likely to correlate with good data.
- Only one tissue section should be placed per slide!
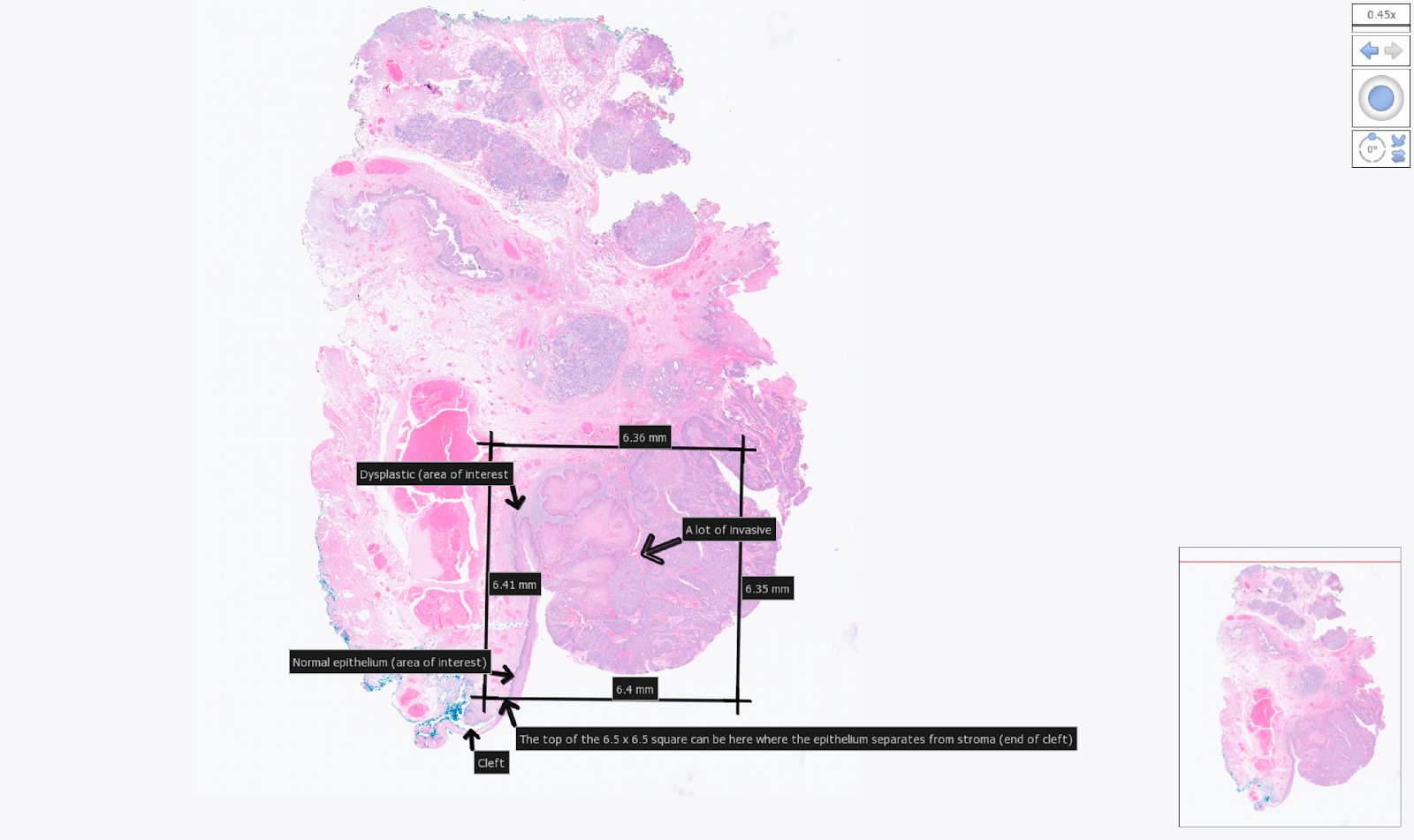
Example tissue section placement for a 6.5 mm CytAssist. Areas of interest within the section were indicated by the user with arrows.
Plan of action
- There are two types of Visium slides with different sizes of the capture areas. The slide type needs to decided upon prior to setting up the project since it will determine the total number of sections used in the experiment:
- 6.5 mm x 6.5 mm (1 kit contains a total of 4 capture areas)
- 11 mm x 11 mm (1 kit contains a total of 2 capture areas)
- We require you to section and place the respective FF sections on coated or charged glass slides e.g VWR SuperFrost Plus Slides.
- Place one section per slide.
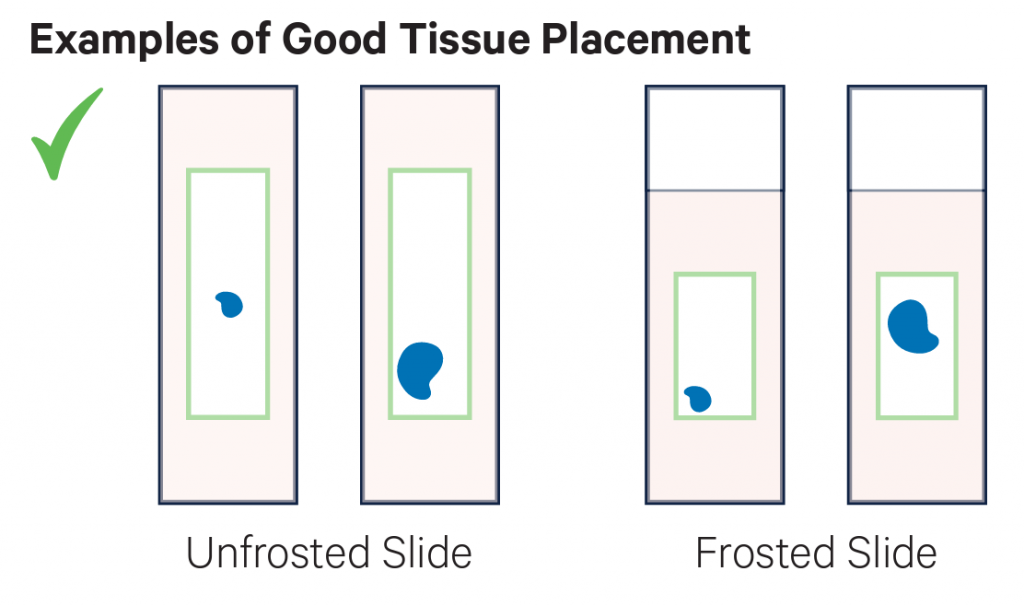
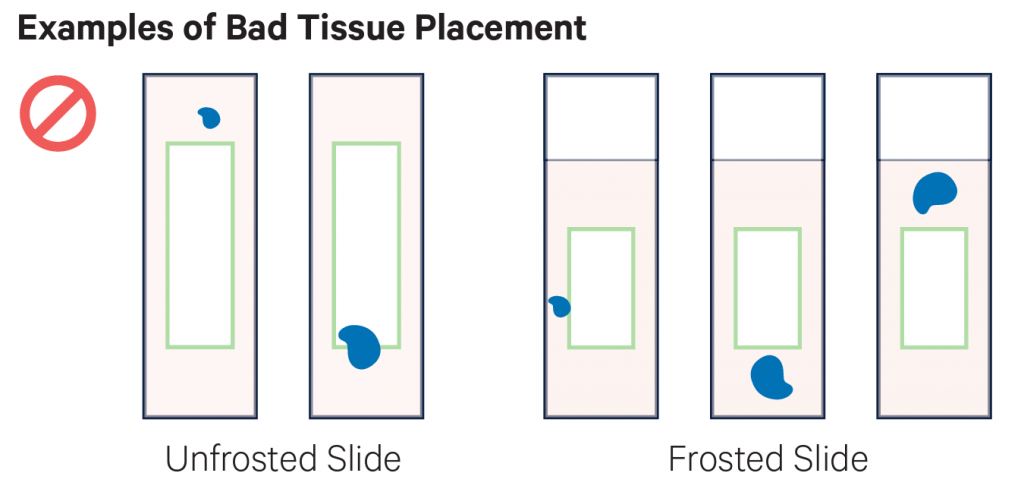
- Sections must be placed within the specific margins described here – sections outside the margins cannot be successfully transferred. To do so, create a representative allowable area on a 75 x 25 x 1 mm blank slide (see image below). Optimise section quality and practice section placement within the allowable area before working with experimental blocks.
- To ensure that the section is in the correct area, we suggest using this printable template.
- The sections placed within the given margins may be larger than the capture areas (see sizes above). Be aware that the area that will be transferred is a square with its edges parallel to the edges of the slide according to the given sizes and the tissue outside the defined areas will not be transferred onto the Visium slides.
- We recommend that you provide us with a back-up sample for each sample that is submitted to us. These will be returned to you upon request.
- Sections must be between 10-20 µm thick. We most often work with sections that are 10 µm thick (refer to this list of FF tissues tested by 10X Genomics for more information).
- After sectioning, keep the slides frozen at all times. Store them at -80ºC in sealed slide mailers until shipment on dry ice.
- Library preparation must be started within 6 months after sectioning and please only section your tissue after consulting with us.
- Check the 10X Genomics website for further information about Visium CytAssist Spatial Gene Expression Fresh Frozen Tissue Preparation and Fixed Frozen Tissue Preparation
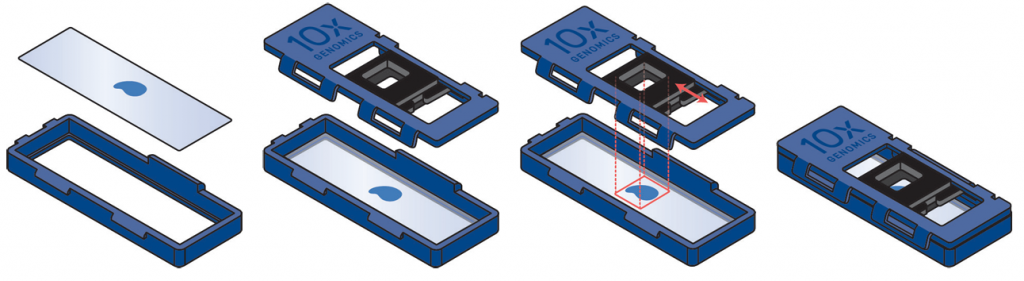
Examples of placement of sections. On the top row there examples of good placement of sections within the margins of the slides. In the middle row there are examples of tissues placed partially or totally outside the margins, and therefore they cannot be used. The bottom row shows the cassette used to transfer the probes to the Visium slide.
Information we need prior to sample delivery:
- Once your order has been accepted, you will receive a project Agreement. Please read it to make sure it reflects your specifications and if everything looks like it should please sign it and return a copy to us electronically.
- A filled-in safety declaration that is sent to you along with the user agreement.
- A filled-in Sample Information Sheet that clearly indicates the sample names and the corresponding RIN scores derived from sections you have analysed.
- Microscope images of H&E stained sections adjacent to the samples, as previously described, clearly highlighting the area of interest (see example above).
- We will send you slide containers with the specific barcodes for the project, which should be used to send the slides to us.
Contact us prior to sectioning to coordinate timing and ensure lab availability so that library preparation can be started within the recommended 6 months from sectioning.
Instructions for sample labelling and delivery
Use the pre-labelled slide containers to send us the superfrost slides with your sections – we issue one container per sample that is ordered. Please place your slide with the section of interest and one backup slide in the same slide container for each sample.
The project-specific barcode label is a combination of your project ID (Pxxxxx) – see user agreement – and a serial number: e.g. `PxxxxxP1` for the first container. Each slide needs to be labelled with the NGI sample IDs from the Visium sample information sheet using stickers or markers, e.g.:
- Container `PxxxxxP1` will contain slides labelled `Pxxxxx_101` and a back-up slide.
- Container `PxxxxxP2` will contain slides labelled `Pxxxxx_201 and a back-up slide.
For shipment, seal properly the slide containers with parafilm and place them in a tightly sealed container/box to limit exposure and keep cold. Send the box in a styrofoam box with sufficient dry ice to prevent the sections on the slides from melting, accounting for transit and delivery times.

Example of slide container. The image shows the slide container that we will be send to the users. It has a label with the barcode corresponding to the project. Inside, they can fit a maximum of 4 slides and which should be individually labelled with the name of the sample.
Summary of the Procedures at NGI
Tissue sectioning is not included in our services using the CytAssist protocol
- H&E staining and imaging
- Transfer of RNA-ligated probes to Visium slides using CytAssist
- Library preparation
- Sequencing
- QC (and analysis with Space Ranger)
Sample and library preparation
Fixation, staining and imaging of the sections is followed by probe hybridisation. The hybridised probes are then transferred to the Visium slide using the CytAssist instrument. Probe extension with integration of spatial barcodes is then performed on the slide, and the subsequent steps are carried out in reaction tubes.
Library QC and sequencing
All individual libraries are assessed for their quality, this includes concentration measurements and estimation of average fragment lengths. Based on these measurements, libraries are pooled and later sequenced on Illumina systems such as the NextSeq or NovaSeq, depending on the sequencing depth required.
A general recommendation is a minimum of 25 000 read-pairs per spot covered by tissue (e.g. if the tissue covers the whole capture area with 5 000 spots, about 125 M read-pairs per section is recommended). Thus, we may need to adjust the sequencing setup after we know the actual tissue coverage and result of library QC. Libraries are sequenced with a read setup with 28 cycles for read 1, two index reads of 10 cycles each and 50 cycles for read 2.
Bioinformatics
Sequencing data QC is handled by the NGI and run through the 10x Genomics Space Ranger analysis pipeline. Raw data and Loupe visualisation files are delivered. You can read more about Spatial Transcriptomics analysis here.
Policy for damaged and/or misplaced sections
NGI will proceed with library preparation unless we deem user-supplied sections to be damaged, sub-optimally placed on slides, or not at a suitable thickness for the method. Therefore, it is in the best interest of users to ensure that our stated sample requirements are strictly adhered to.
Signing an NGI-issued agreement for your project signifies that you accept that our staff can decide how to proceed with samples that are submitted to us. Decisions may include but are not limited to:
- Proceeding with samples that do not strictly fulfill our sample requirements but have a fair chance of yielding libraries (albeit at a lower quality than would otherwise be expected), etc.
- Charging a flat fee of SEK 5000 as a processing fee for every back-up sample that is processed at NGI if the user has not adhered to our sample requirements for section placement on slides.
- Aborting projects comprising samples that are suboptimal for Visium.
Note that we can never guarantee any outcomes of library preparation or the preceding steps for the Visium protocol, especially if samples do not meet the criteria defined by 10X Genomics and that we outline.
Last Updated: 1st July 2024