Olink Explore 384-3072 multiplex protein analysis
Method for multiplex protein biomarker analysis with sequencing readout using the Illumina NovaSeq platform.
As a joint effort between the SciLifeLab National Genomics Infrastructure (NGI) and the Affinity Proteomics Uppsala unit (APU) at SciLifeLab Clinical Proteomics and Immunology platform, the Olink Explore assays are available in our services. Olink Explore is based on the Proximity Extension Assay (PEA) but unlike the qPCR 24 to 92-plex Target-line, the Explore assay utilizes high throughput sequencing. The shift in readout technology has increased the multiplexing capacity and with the largest panel available today over 3000 proteins are analyzed in parallel for each sample.
Below you can find some basic information regarding samples, panels and data output. If you have any further questions, please contact us at explorelab@scilifelab.uu.se for more information about our service, or visit Olink´s website, Olink Explore – Olink
Sample Requirements
- Sample type: Plasma, serum or CSF. NB! All samples in a project must be of the same type, even the same plasma type is required. It is possible to combine plasma and serum on the same sample plate for analysis but CSF must be analyzed separately.
- Number of samples: 88 samples is the lowest number of samples allowed.
- Volume of sample needed:
- >30-40 µL if up to 4 x 384 Explore panels to be analyzed
- >60-80 µL if Explore 3072 panel to be analyzed
- Plate design:
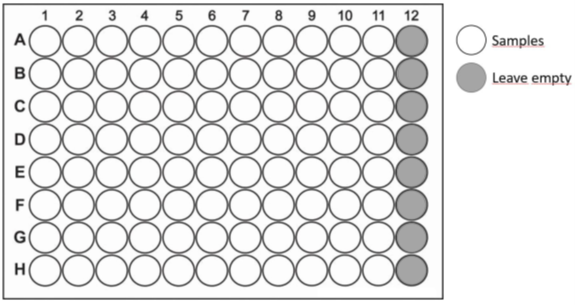
Each plate is designed to include 88 samples and 8 controls. The samples should be placed in the wells A1-H11 according to the template below. The last column (A12-H12) is reserved for the controls and must be empty. All controls are provided by the Facility.
Please make sure to use a 96-well plate and sealer that are suitable for storage at -80°C. The plate should preferably have V-shaped wells. We recommend the ABgene, AB-0800-L plate and ABgene, AB-0558 sealer. Plates and sealers can be provided upon request.
- Randomization:
Samples must be randomized within and between the plates included in the project. This in order to minimize the risk of removing true biological differences during QC and to be able to check for row and column effects.Sample Requirements
Available panels
Eight separate 384-plex protein panels are available and can be ordered separately, or combined to form the Explore 1536 or Explore 3072 assay format. The panels are:
- Olink Explore 384 Cardiometabolic I and II
- Olink Explre 384 Neurology I and II
- Olink Explore 384 Inflammation I and II
- Olink Explore 384 Oncology I and II
A set of twelve protein control assays are included in each 384-panel, and eight control samples are included in every sample plate to monitor the assay performance and the quality of the data generated. The controls are also used for the normalization during data processing.
Description of the method
The PEA technology is based on two matching antibodies binding to a protein, upon binding, the DNA tags from the two antibodies come into contact/proximity and the DNA tags hybridize and are extended making a unique DNA barcode for each protein analyzed. After amplification, digital DNA counting is performed using sequencing and the resulting counts directly relate back to the concentration of the protein in the sample. We evaluate the quality of the sequencing library prior to sequencing. The sequencing will be carried out the specific setup for the application as stated in the agreement.
Data processing, quality control and delivery
During the sequencing run, known sequences are read and the BCL files generated are subsequently converted to counts files. The counts directly represent the protein concentration in the sample analyzed.
The Counts file is processed in Olink proprietary software MyData to generate NPX (Normalized Protein eXpression) values. During this process and extensive quality control is performed where the control samples and markers are evaluated to rule out bad assay performance and variation between plates. The quality control samples are also used for normalization. Data from the Olink Explore assay is delivered as NPX values and accompanied by a Certificate of Analysis describing the project and data. The data is delivered encrypted using CryptShare.
Last Updated: 6th October 2022