Spatial transcriptomics (Visium)
Spatial transcriptomics allows you to map gene expression across intact tissue sections, combining histological context with transcriptome-wide profiling using 10x Genomics Visium platforms for fresh frozen and FFPE samples
Spatial transcriptomics enables the study of gene expression in intact tissue sections while preserving spatial information. This technology combines histological context with transcriptome-wide profiling, allowing researchers to map cell types, states, and interactions directly onto tissue architecture.
At NGI we provide support for several spatial transcriptomics approaches, primarily based on the 10x Genomics Visium platforms. These methods allow you to capture polyadenylated RNAs from fresh frozen tissues and use probes covering the whole transcriptome on FFPE tissues to generate high-resolution maps of gene activity. Our service includes guidance on sample preparation, library construction, sequencing, and basic downstream analysis.
Explore our current spatial transcriptomics applications below to find the approach that best suits your project. Please feel free to contact us contact us for advice on experimental design, tissue requirements, or other questions.
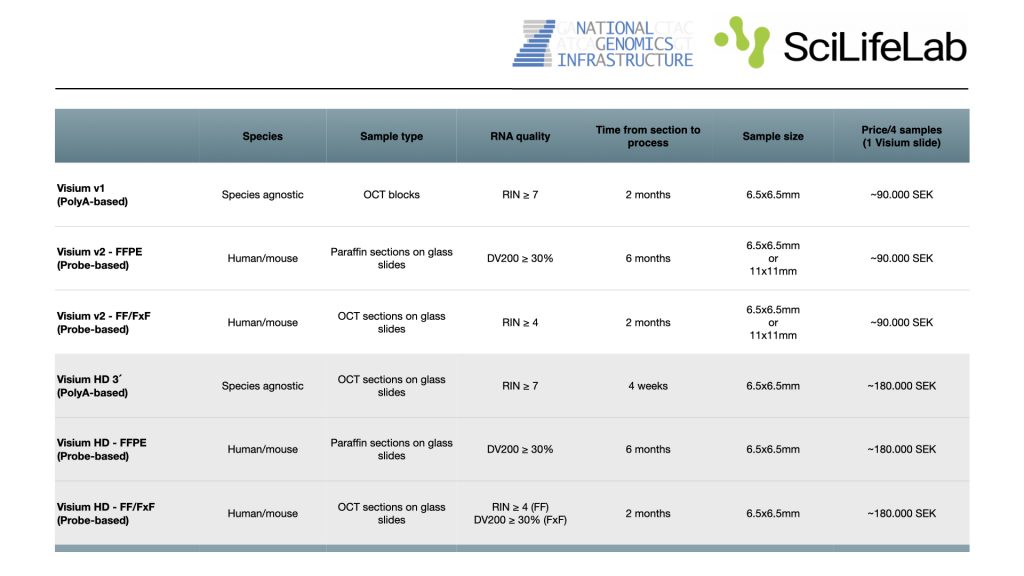
Visium HD 3´ – PolyA-based
NGI offers high resolution spatial transcriptomics through the 10x Genomics Visium HD 3’, which combines histology with PolyA-based transcriptomics in HD spatial context
spatial transcriptomics RNA illuminaVisium HD – Probe-based – FF/FxF samples
We now offer high resolution spatially resolved transcriptomics through the 10X Genomics Visium HD CytAssist, which combines histology with probe-based transcriptomics in HD spatial context
transcriptomics RNA illumina spatialVisium HD – Probe-based – FFPE samples
We now offer high resolution spatially resolved transcriptomics through the 10X Genomics Visium HD CytAssist, which combines histology with probe-based transcriptomics in HD spatial context
transcriptomics RNA illumina spatialVisium – Probe-based – FF/FxF samples
We now offer spatially resolved transcriptomics through the 10X Genomics Visium CytAssist, which combines histology with probe-based transcriptomics in a spatial context.
transcriptomics RNA illumina spatialVisium – Probe-based – FFPE samples
We now offer spatially resolved transcriptomics through the 10X Genomics Visium CytAssist, which combines histology with probe-based transcriptomics in a spatial context.
illumina spatial transcriptomics RNAVisium 3´- PolyA-based
We offer spatial transcriptomics through the 10X Genomics Visium method. The method combines histology with unbiased transcriptomics in a spatial context.
transcriptomics RNA illumina spatialSpatial Transcriptomics analysis
Demultiplexing, image alignment, read alignment and barcode/UMI counting of sequencing reads generated by the 10X Genomics Visium assay.
transcriptomics RNA illumina spatial