Single-Cell Multiomics with BD Rhapsody HT Xpress
BD Multiomics Award: NGI together with BD Biosciences currently offer an award for one researcher to run 4 samples of whole transcriptome analysis. For more information on what is included and how to apply see here.
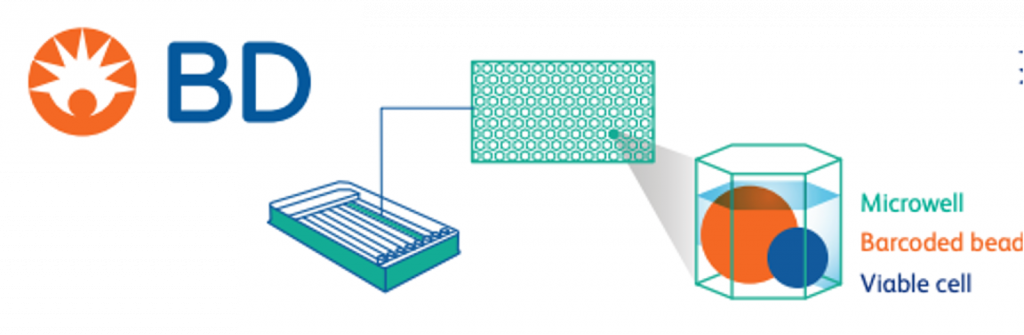
BD Rhapsody HT Xpress offers single-cell analysis of multiple modalities. Single-cell status is achieved through a combination of microwells and barcoded beads for nucleic acid capture. Barcoded beads introduce both cell-specific barcodes and UMIs during reverse transcription or amplification.
Once RNA is captured on the beads and converted to cDNA, the sample is stable at 4°C and beads/cells can be subsampled if desired.
Whole transcriptome analysis
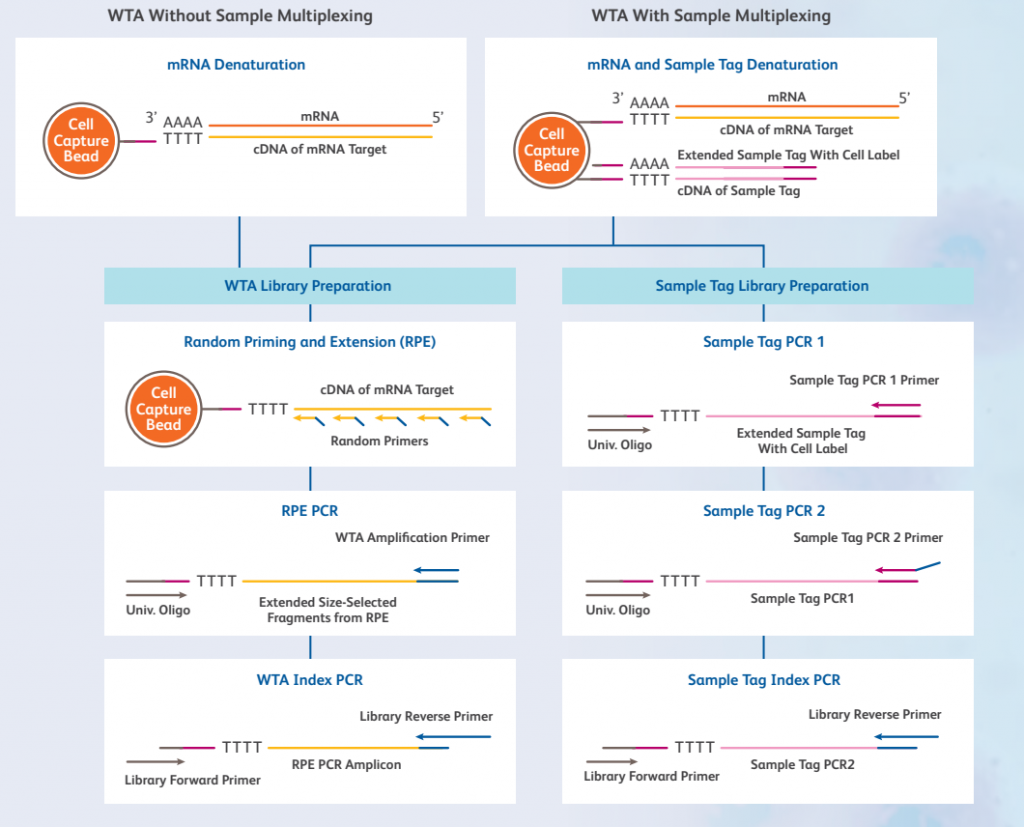
For whole transcriptome amplification analysis, RNA is captured on the beads by poly-dT oligos and used to prime first strand synthesis in reverse transcription. The resulting cDNA is attached to the beads and can be stored at 4°C for up to a year before proceeding with library preparation.
Next, the cDNA is used for Random Priming and Extension (RPE) before amplification and finally indexing in Library construction. Compatible with sample multiplexing, TCR/BCR analysis, and CITE-seq.
Compatible sample types
The Rhapsody workflow is compatible with cell (gene expression experiments) or nuclei (ATAC and Multiome experiments) suspensions from diverse species.
Sample requirements
In order to obtain high quality data from the experiment the sample should have:
- No or minimal amounts of cell debris.
- No or low levels of cell/nuclei aggregates.
- Cells quality:
- High cell viability. We recommend >90% viability if possible to ensure high data quality. BD does not recommend using suspensions <50% viable.
- Cell size <20uM in diameter, larger cells may reduce capture efficiency.
- Nuclei quality
- High quality with intact nuclear membrane.
- Few intact cells
We always recommend performing a trial prep for any new sample types before handing over any samples to NGI. The sample quality should be assessed before sample delivery. For nuclei, we recommend to assess nuclei quality at 40x or 60x magnification in a microscope.
Extra options
The Rhapsody workflow is compatible with a wide repertoire of multiomic assays/options, in addition to the whole transcriptome assay described above, such as:
- TCR/BCR analysis
- CITE-seq
- ATAC
- Multiome (ATAC and GE)
- Cell hashing using antibodies
- Targeted RNA-seq
Bioinformatics
Basic bioinformatic analysis using the BD Rhapsody pipeline. Your data will be delivered to you through the DDS online delivery system
Last Updated: 3rd February 2026