Visium 3´- PolyA-based
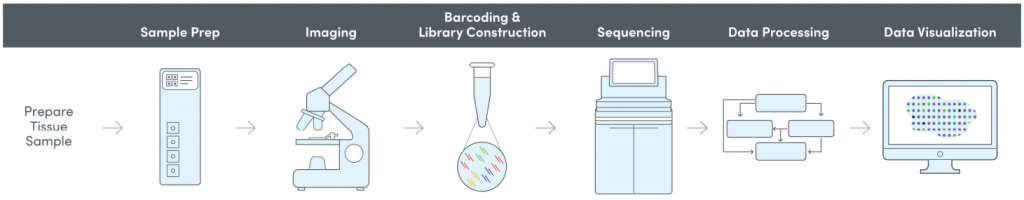
We offer spatial transcriptomics through the 10X Genomics Visium method. The method combines histology with unbiased transcriptomics in a spatial context.
Allowing visualisation of the dynamics of transcription directly on top of tissue morphology gives unprecedented insight into developmental and pathological processes. NGI offers Spatial Transcriptomics as a service, using the 10X Genomics Visium Spatial Gene Expression for Fresh Frozen.
For more info, please refer to our online webinar on Spatial Transcriptomics 10X Visium.
Please contact us for a discussion prior to planning your experiments.
Sample requirements
Fresh frozen OCT embedded tissue, with intact morphology and high RNA quality.
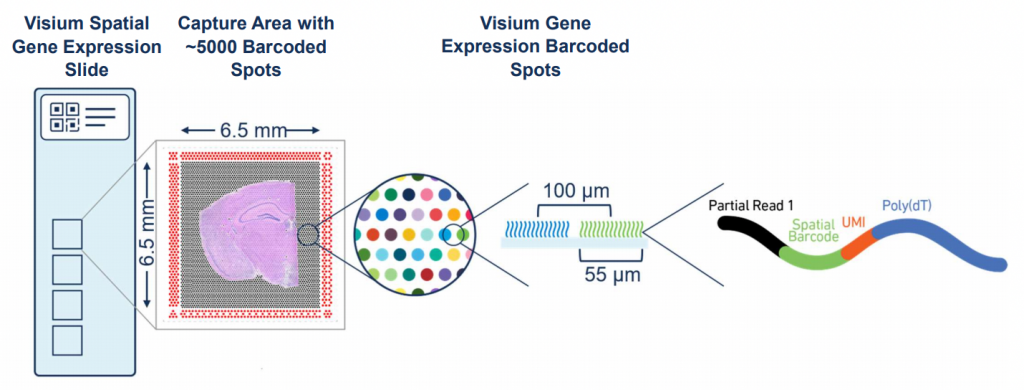
Please keep in mind that the maximum size of the tissue block has to be no more than 6x6mm (not larger than the predefined capture area of the array).
Any staining optimisations need to be done by the user otherwise we will follow 10X Genomics’ standard protocol.
How to evaluate the sample quality
We require our users to do their own QC steps before sending samples. For this library prep method, we require:
- RIN value > 7 (if possible)
- RNA extraction and RIN value control on a Bioanalyzer or similar, preferably on the sample you will use.
- RIN value above 7 is not always possible, please get in touch to discuss if this is a problem.
- H&E staining pictures that you can send to us.
- (Thickness optimisation)
- If possible, a one-cell thick layer
- This typically involves a trade off, most tissues are sectioned at 10 µm
More info on optimised tissue types by 10X Genomics is found here.
If you are not able to carry out these steps, or your samples are below the required thresholds, please get in touch.
Included in the method
- Cryosectioning
- Imaging
- Tissue optimisation (TO)
- Library preparation (LP)
- Sequencing
- QC (and analysis)
Tissue optimisation
This method requires a tissue optimisation (TO) to establish the right permeabilisation conditions for each specific tissue type. This includes cryosectioning, permeabilisation, cDNA synthesis, tissue removal, imaging and evaluation.
Please note that the H&E intensities can differ from slide to slide.
Depending on the nature of your tissue and the number of different tissue types this step may have to be performed on more than one TO slide.
Sample and library preparation
In order to generate the Visium libraries, sections from the OCT embedded tissue are aligned on the capture areas of the Visium gene expression slides (4 areas per slide, NGI will process whole slides only, so you can only order multiples of 4 sections).
Sections are permeabilized according to the results of the tissue optimization after which one library per capture area is prepared (total of 4 libraries per gene expression slide).
Library QC and sequencing
In this step, we evaluate the yield obtained and we will also determine the size distribution of the libraries generated. We will inform you of the QC status of each sample. Once the libraries have passed this QC step, they are queued for sequencing.
Visium libraries are typically sequenced on Illumina systems such as the NextSeq and NovaSeq, depending on the sequencing depth required. A general recommendation is ca 50 000 read-pairs per spot covered by tissue (e.g. if the tissue covers the whole capture area with 5 000 spots, about 250M read-pairs per section is recommended). The read setup used is 28-10-10-90.
Bioinformatics
Sequencing data QC is handled by the NGI and run through the 10x Genomics Space Ranger analysis pipeline. Raw data and Loupe visualisation files are delivered. You can read more about Spatial Transcriptomics analysis here.
General FAQs
How do I fresh freeze and embed my tissue in OCT?
Tissue has to be snap-frozen in cold isopentane (not liquid nitrogen). More info is found here and instruction videos are found here and here. Preferably snap-freezing and embedding is performed at the same time but can be performed in separate steps.
What tissue types are supported?
A list of different tissue types supported by 10X Genomics is found here. However, we have successfully prepared sequencing libraries for tissue types not included on their list. Please get in touch with us if you want to discuss your tissue type further. For now, we do not accept bone/cartilage or plant tissue sample.
We lack bioinformatics competence. Can I get any support for that from NGI?
NGI will run the Space Ranger analysis pipeline for you. For further suport on downstream analysis, visualization and integration of spatial datasets we recommend to NBIS: info@nbis.se
NBIS also have a weekly drop in session (Tuesdays 14:00-15:00) where you can ask questions directly: https://nbis.se/events/
We received 10X Visium data and have bioinformatics competence within the group. Can you direct us with suggestions on available tools for downstream data analysis.
You find a few suggestions of available R and Python packages here (scroll down to the bottom of the page).
What does it cost to run a Visium project?
Please refer to the 10X Visium cost examples here (click on the “Illumina Sequencing” tab and scroll down to “Cost for Spatial Transcriptomics 10X Visium”)
Last Updated: 23rd December 2025