10X Genomics Multiome (ATAC + gene expression)
Profiling of 3´gene expression and chromatin accessibility in the same cell.
We offer single cell transcriptome and chromatin accessibility profiling using the 10X Genomics Multiome kit. The Multiome kit combines single nuclei RNA-seq with single cell Assay for Transposase-Accessible Chromatin using sequencing (ATAC-seq) within the same cell. Using the 10X Genomics Multiome kit, the nuclei are transposed in a bulk solution and subsequently partitioned into a Gel Beads-in-emulsion (GEMs) using a microfluidic chip. Employing a pool of approximately 750,000 10x Barcodes, both transposed DNA and cDNA are uniquely indexed. Following library generation and sequencing, the 10x Barcode is used to assign individual reads to individual nuclei.
Compatible sample types
The 10X Multiome kit is only compatible with nuclei suspensions.
Sample requirements
In order to obtain high quality data from the experiment the sample should have:
- No observable debris or aggregation.
- No inhibitors of reverse transcription or GEM generation.
- A viability of the cell suspension of more than 90% (before nuclei isolation)
- Intact nuclei of more than 90%
- Intact nuclei with high-quality nuclear membranes and less than 5% of intact cells. (nuclear membranes are well resolved and nuclei show no visible sign of blebbing)
- A concentration of 160-8,060 nuclei/μl (concentration depends on targeted nuclei recovery; see table in user guidelines)
- Use only RNase inhibitors that are recommended by 10X Genomics!
We recommend performing a trial prep and assessing nuclei quality at 60x in a microscope. The quality of the nuclei suspension to be used with the Multiome kit plays a crucial role in obtaining high quality sequencing data of captured nuclei. Please visit 10X Genomics webpage for “Demonstrated Protocols’‘ for guidance on how to obtain high quality single nuclei suspensions.
Starting a Multiome project with NGI
- Contact us at support@ngisweden.se to discuss your project.
- Read our current user guidelines here (v5).
- Submit a project request through the iLAB system (search for the ESCG facility). A project coordinator will set up your project and get back to you. Please check the project setup to make sure it reflects your specifications and if everything looks correct, agree to the terms and conditions within iLAB.
- If possible – run a pilot for the sample preparation to ensure that it yields high quality cell suspensions/nuclei.
- Book a day/days to bring your samples to us.
Information we need prior to sample delivery
- An electronically accepted project request in iLAB, with no remaining questions about setup or number of samples.
- A filled-in safety declaration that is sent to you along with the user agreement (if applicable).
Sample processing at NGI
Upon delivery of the samples to NGI, the responsible lab staff will count the nuclei and check that the nuclei suspension is free of aggregates and debris before you leave. It is your responsibility to ensure that the nuclei are of high-quality.
Should the sample be sub-optimal and you still wish to continue with the experiment, we will ask you to confirm this in writing. In such a situation you will be charged the cost of the 10x reactions regardless of the outcome. If you decide not to continue, there will be no charge unless a kit was purchased specifically for you (communicated to you before setting up your project).
All sample manipulation such as labeling, multiplexing etc must be performed by the researcher prior to delivery of the sample. The delivered sample should be ready to load on the 10X Chromium instrument.
Library preparation
1. Transposition
A transposition mix, containing a transposase, is added to the nuclei suspension. The transposase inserts its adapters preferentially into open chromatin regions which can later be amplified and sequenced.
2. GEM generation and barcoding
The samples are loaded on the Chromium X controller for GEM generation. Fragments containing a transposase-added adapter receive a 10x Barcode and are amplified inside the GEMs. During cDNA synthesis a cell-specific barcode is incorporated into the full-length cDNA inside the GEMs. The barcodes will be used to bioinformatically assign fragments to single nuclei.
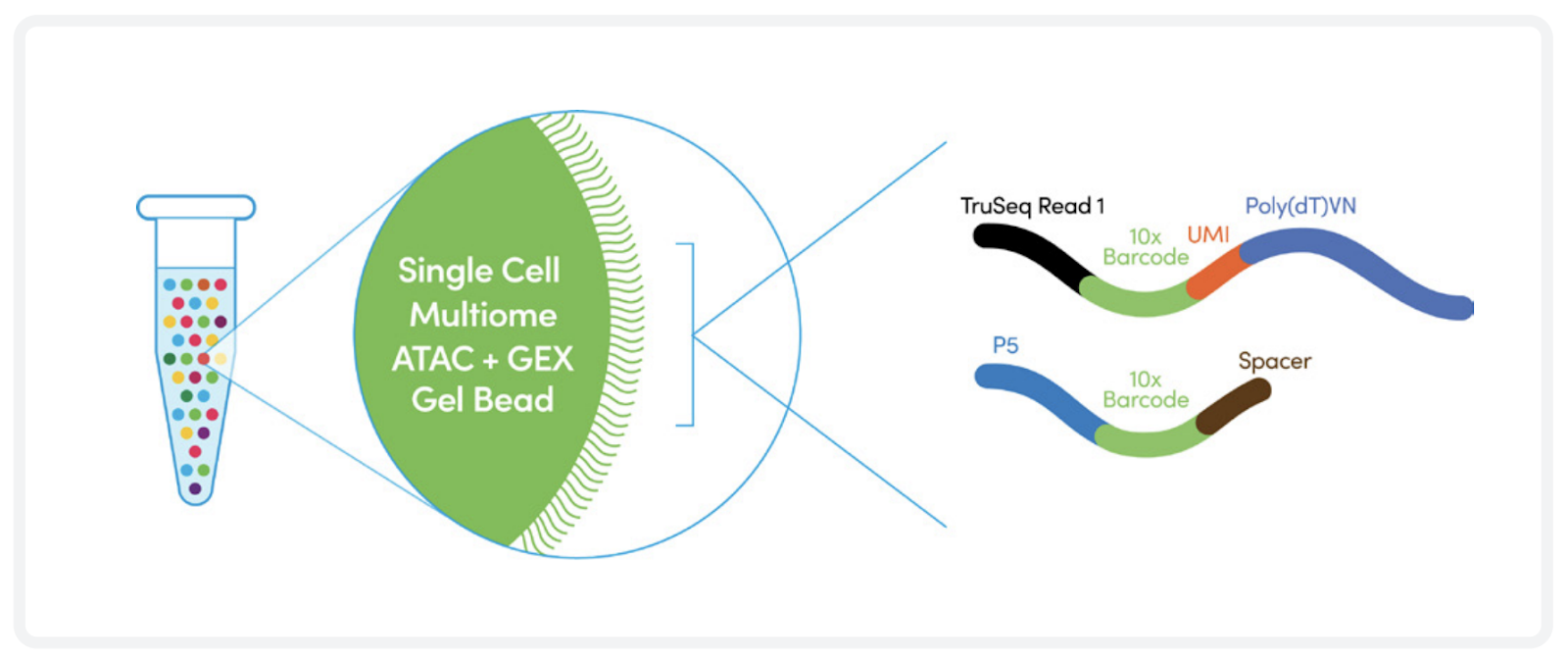
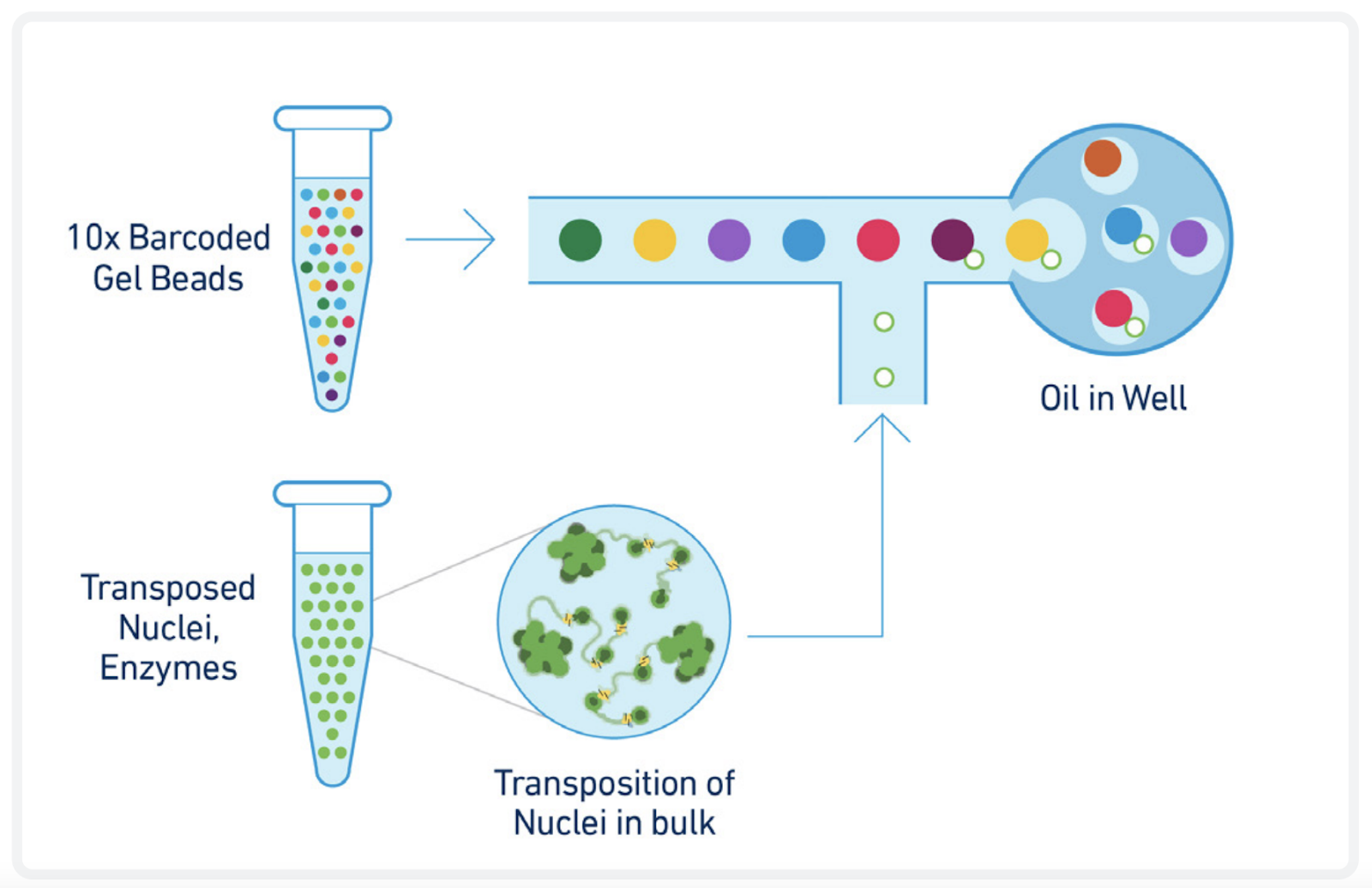
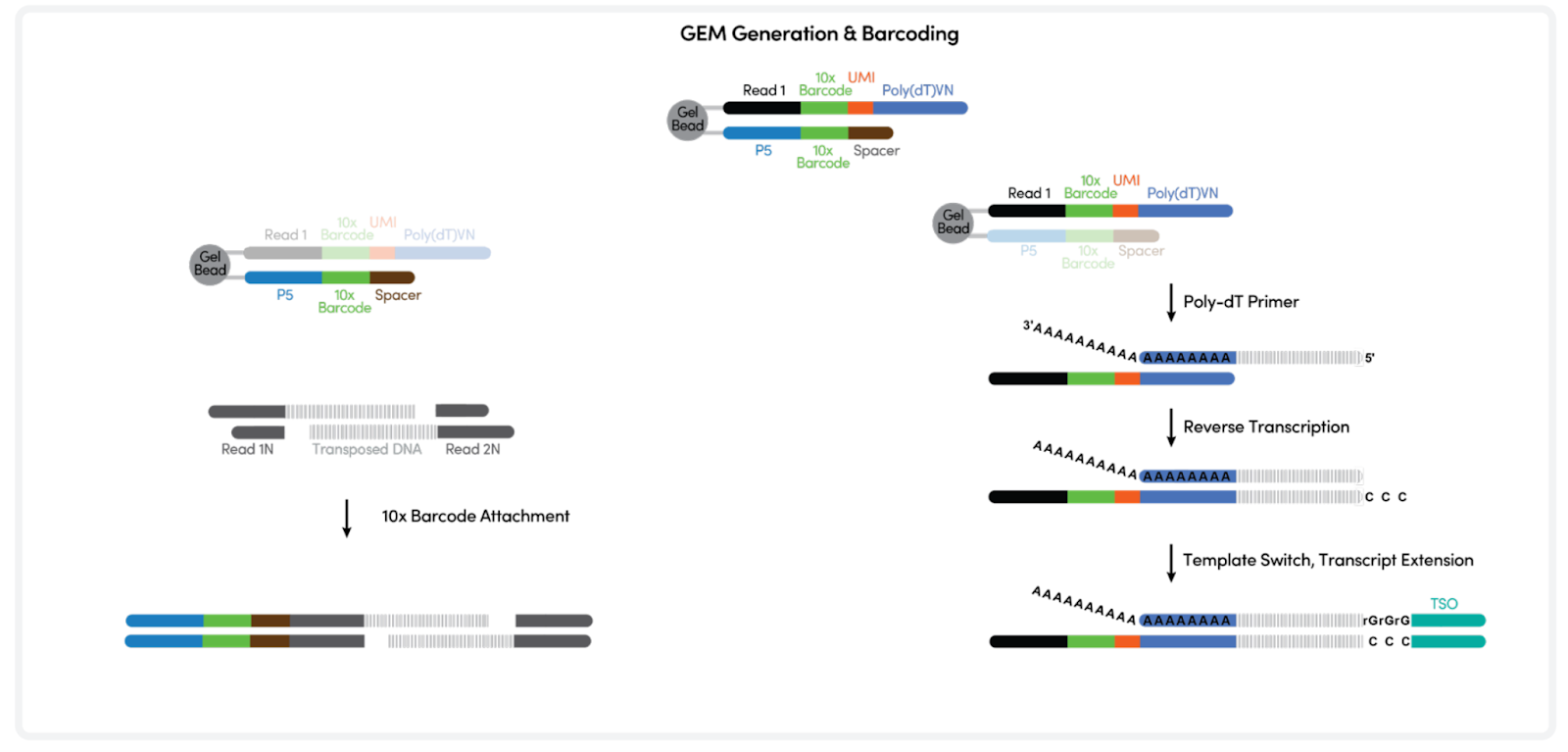
10X Multiome GEM generation (top). 10x barcode incorporation (bottom). Adapted from https://cdn.10xgenomics.com/image/upload/v1666737555/support-documents/CG000338_ChromiumNextGEM_Multiome_ATAC_GEX_User_Guide_RevF.pdf
3. Post-GEM Clean-up & Pre-Amplification PCR
The emulsion is broken by addition of a recovery agent and the amplified fragments are purified, after which a pre-amplification PCR is performed. The product is used for generating both the ATAC library and for cDNA generation in order to make a gene expression (GE) library.

Barcoding and pre-amplification of transposed DNA (left) and cDNA (right). Adapted from https://cdn.10xgenomics.com/image/upload/v1666737555/support-documents/CG000338_ChromiumNextGEM_Multiome_ATAC_GEX_User_Guide_RevF.pdf
4. ATAC Library preparation
Illumina indexes are added during PCR in bulk.
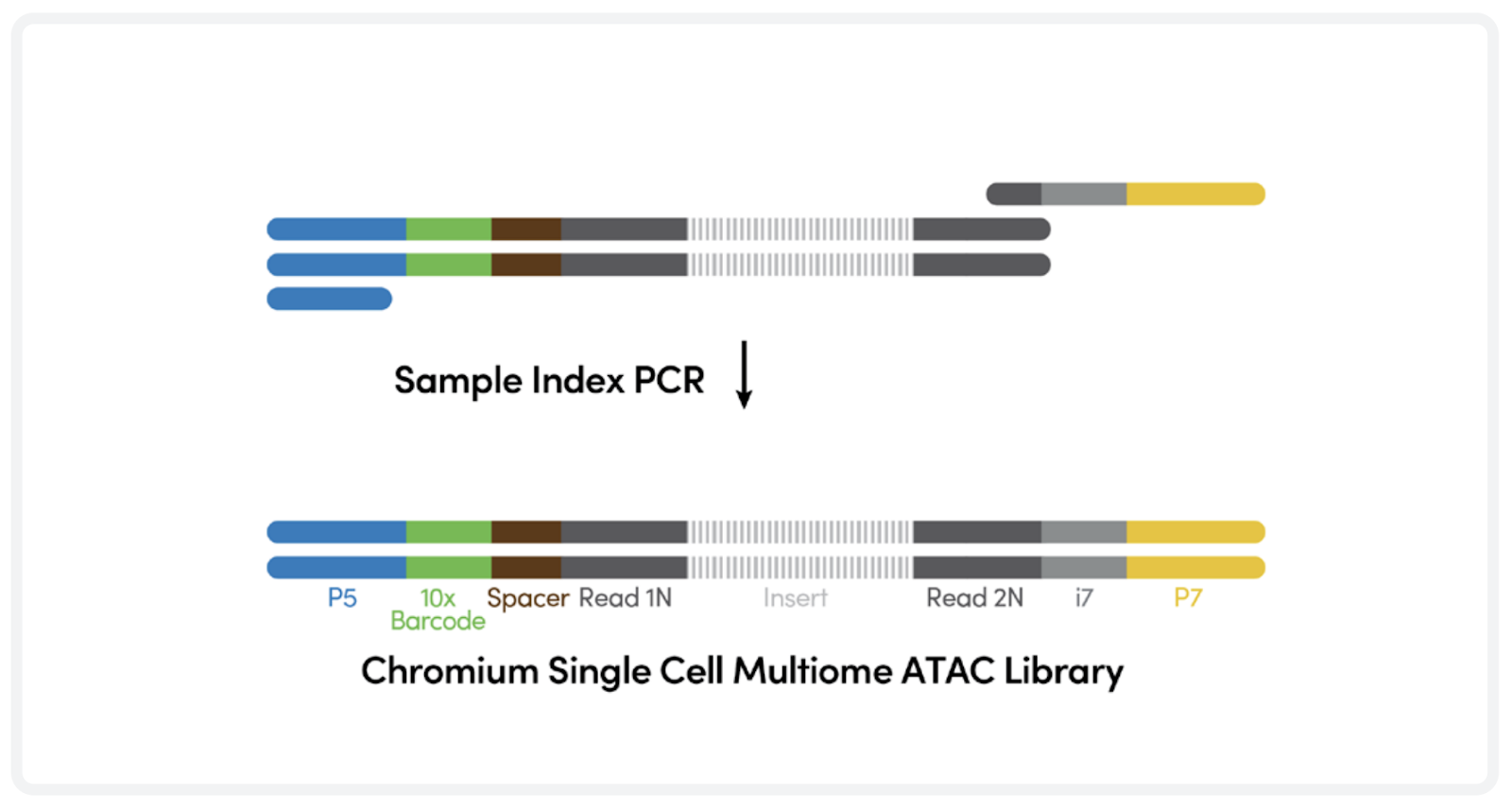
ATAC library construction. Adapted from https://cdn.10xgenomics.com/image/upload/v1666737555/support-documents/CG000338_ChromiumNextGEM_Multiome_ATAC_GEX_User_Guide_RevF.pdf
5. cDNA amplification and library construction
Full-length cDNA is amplified before library construction. Sample Indexes are added in the next step via PCR. The GE libraries get QC-checked through BioAnalyzer, Qubit and qPCR before pooling and sequencing.
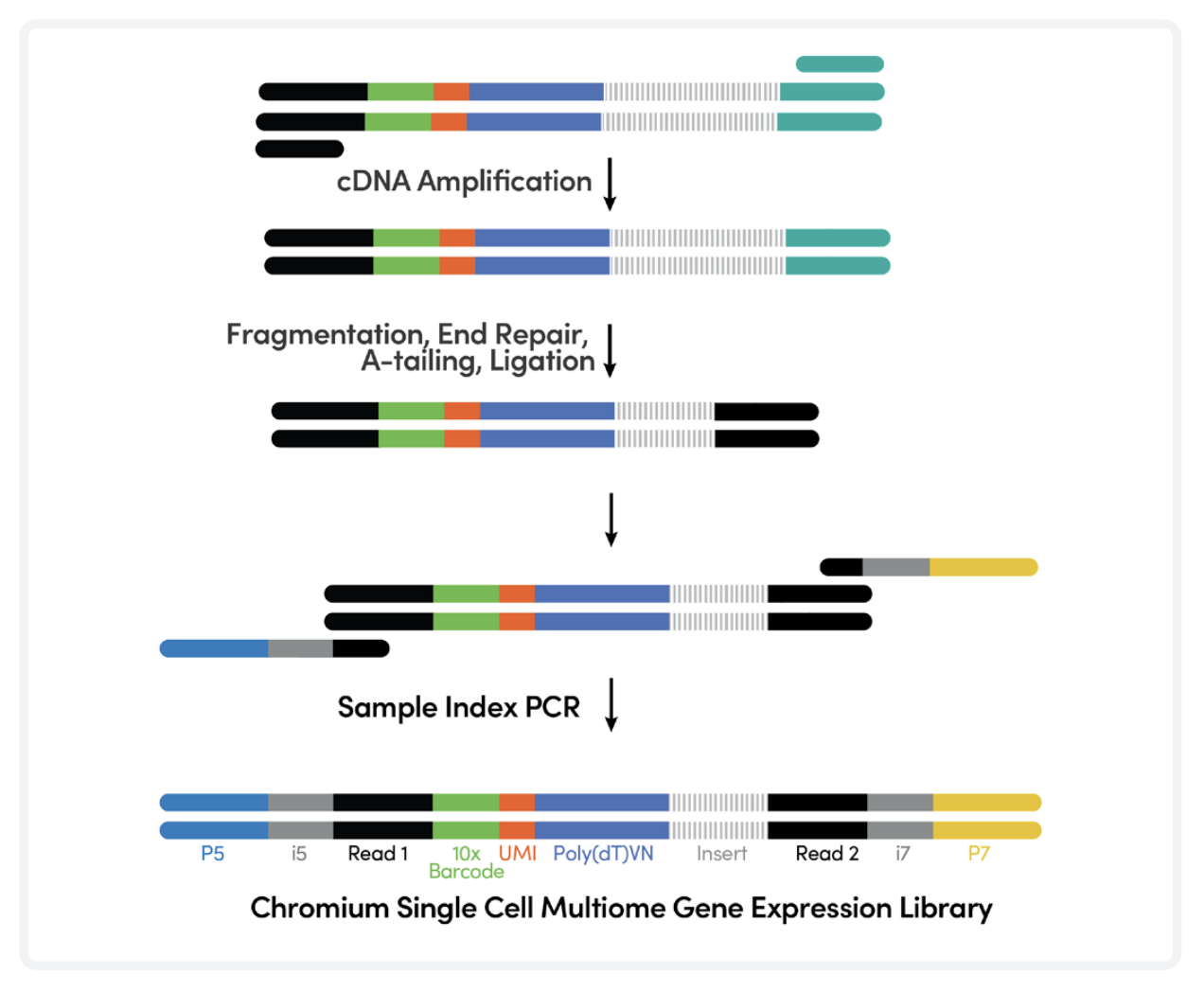
cDNA amplification and library construction. Adapted from https://cdn.10xgenomics.com/image/upload/v1666737555/support-documents/CG000338_ChromiumNextGEM_Multiome_ATAC_GEX_User_Guide_RevF.pdf
6. Library QC and Sequencing
The ATAC and GE libraries are QCed by Qubit, BioAnalyzer and qPCR before pooling for sequencing.
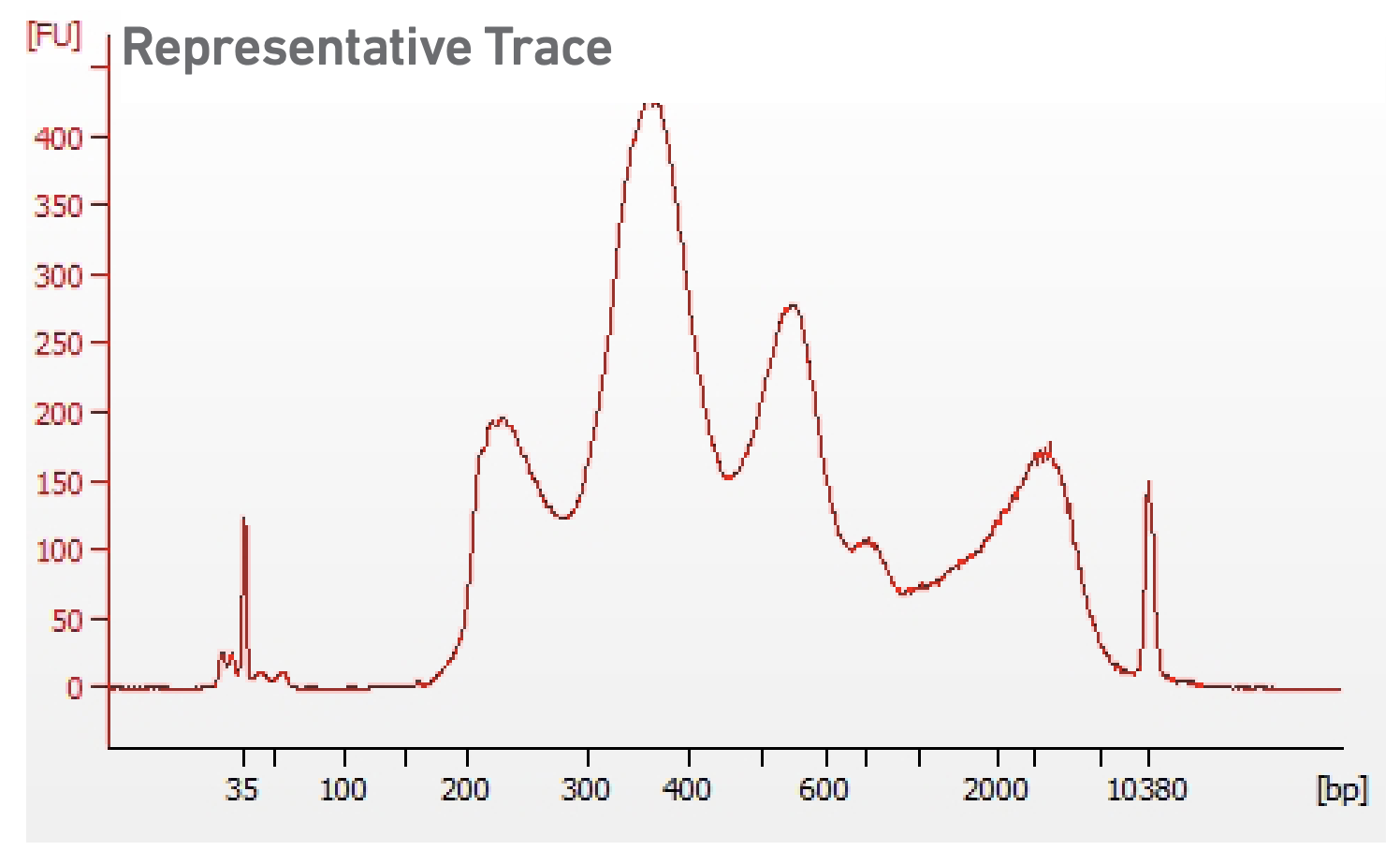
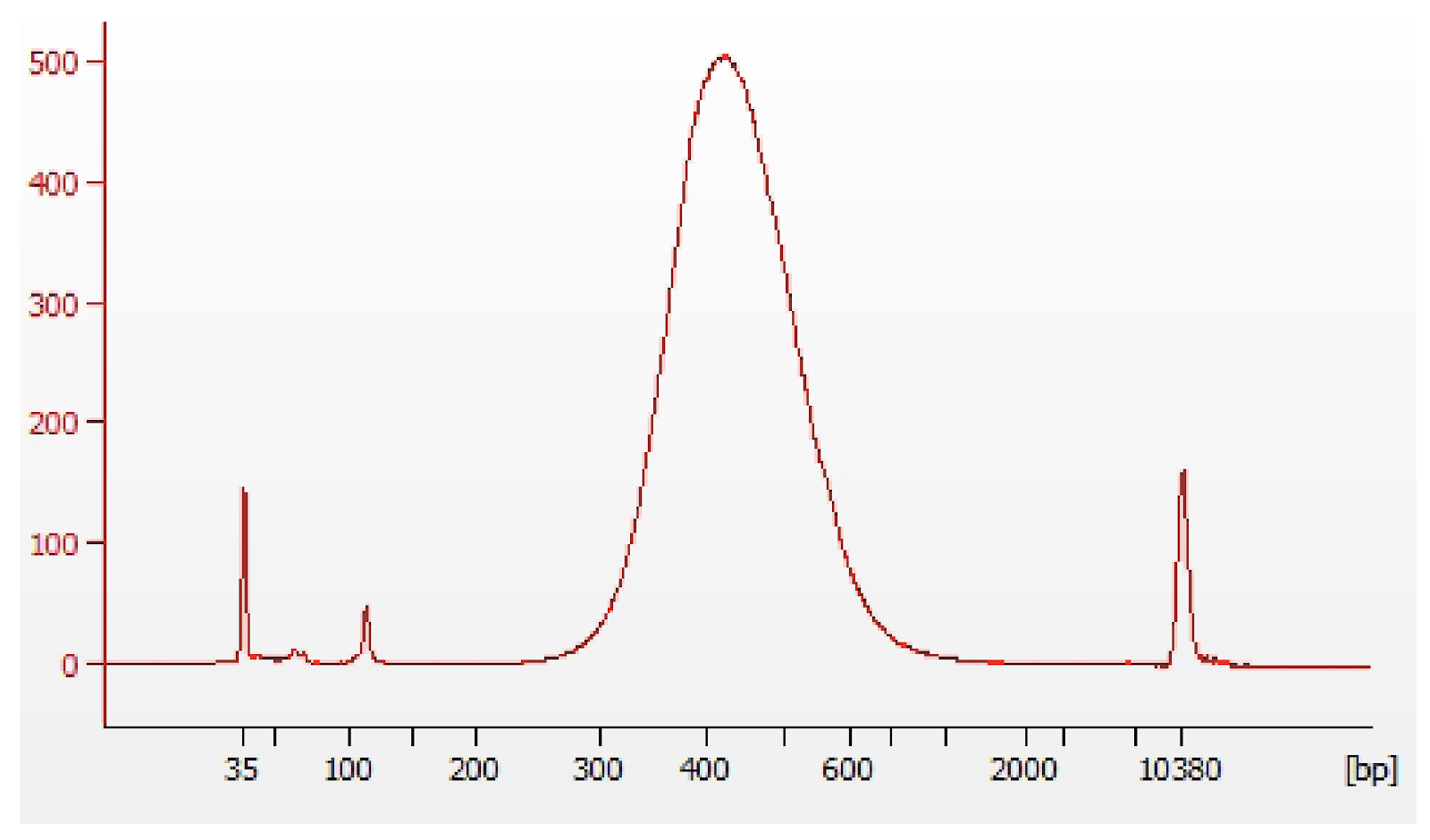
Multiome ATAC (top) and GE (middle) library structure. Representative trace of an Mulitome ATAC (bottom left) and GE (bottom right) library. Adapted from https://cdn.10xgenomics.com/image/upload/v1666737555/support-documents/CG000338_ChromiumNextGEM_Multiome_ATAC_GEX_User_Guide_RevF.pdf
Bioinformatics
Basic bioinformatic analysis using the CellRanger software from 10X Genomics. Your data will be delivered to you through the DDS online delivery system.
Last Updated: 16th May 2024