RAD-sequencing
Genotyping-by-sequencing without prior genome information.
Description
RAD (Restriction-site Associated DNA) sequencing is a method by which certain restriction enzymes are used to fragment genomic DNA samples, followed by size selection and sequencing of molecules within a certain size range. This enables reproducible enrichment of a particular set of genomic loci, where the loci retained for sequencing are more or less randomly distributed over the genome. The sequence data resulting from this application may be used for downstream applications including, but not limited to: Linkage analysis, inference of population genetics metrics and polymorphism discovery.
Sample requirements
We run RAD-seq library preps in batches of 94 samples. We use two controls in worksets, please consult our sample submission page for details on how to load your samples in plates.
| Input | Purified EcoRI-cut DNA. Gel results must be provided. |
| Amount | >200 ng |
| Concentration | 16–100 ng/µL |
| Volume | 16–170 µL. Approx 2 μL of the sample will be used for our initial quality checks, so please account for this when sending us samples. |
| Sample Buffer | 10 mM Tris, pH 8–8.5 or similar (e.g. Qiagen’s EB). |
How to evaluate the sample quality
We check your samples upon arrival, however we still require our users to do their own QC steps before sending samples. For this library prep method, we require the following to be included when submitting sample information.
Making sure concentration, volume and amount fulfil the criteria listed in the table above (this applies to measurements AFTER restriction digest and inactivation of the enzyme).
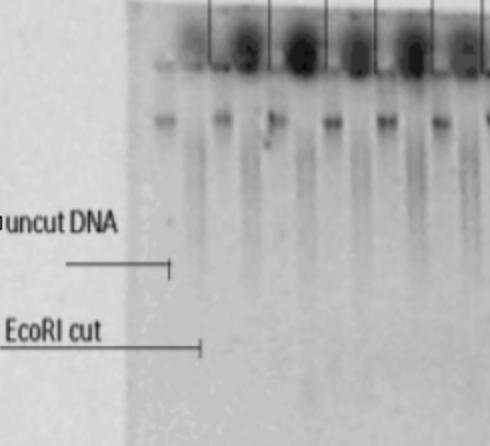
The EcoRI-cut DNA should be run on a gel with digested/undigested samples side-by-side, to confirm successful digestion.
If you are not able to carry out these steps, or your samples are below the required thresholds, please get in touch.
What we do with your samples
EcoRI-digested samples are prepared using the Agilent Bravo system (protocol available at Github: https://github.com/ngi-automation/rad-seq).
Library preparation
During preparation a custom adapter with complementary bases to the EcoRI overhang is ligated to the ends of the digested DNA using T4 DNA ligase and 10X T4 DNA ligase (New England Biolabs). The ligated adapter contains the partial Illumina adapter. Samples are purified using AMPure XP bead purification (Beckman Coulter), strand displacement is performed using Bst 2.0 polymerase, 10X ThermoPol buffer (New England Biolabs) and dNTPs (Thermo Fisher Scientific), another bead purification follows before amplification and indexing by PCR. Finally, the libraries are cleaned up in another bead purification.
Library QC and sequencing
To assess the quality of the library, the concentration is measured with the Qubit dsDNA HS assay (Thermo Fisher Scientific) and the fragment size distribution is checked using the Fragment Analyzer (Agilent). A calculated concentration in the range of 420–720 bp is used to even out the different samples contribution to the pool before subjecting the pool to size selection with the BluePippin (Sage Science). Concentration and size of the pool is analyzed with Qubit and Fragment Analyzer (or Bioanalyzer) before sequencing according to agreed upon setup.
Expected results
It is expected that the resulting sequence data from each sample is enriched for loci in the genome that were generated by EcoRI cuts and that the libraries from each sample will feature mostly the same sequenced loci.
Bioinformatics
Analysis with STACKS: http://catchenlab.life.illinois.edu/stacks/
Last Updated: 22nd January 2026