Illumina 16S sequencing
Sequencing of the 16S gene to study bacterial diversity and composition in a sample.
This application – originally optimised to study the bacterial content of environmental DNA samples – is suited to help identify the presence of multiple bacterial species by sequencing hyper-variable parts of bacterial genes (specifically 16S rDNA).
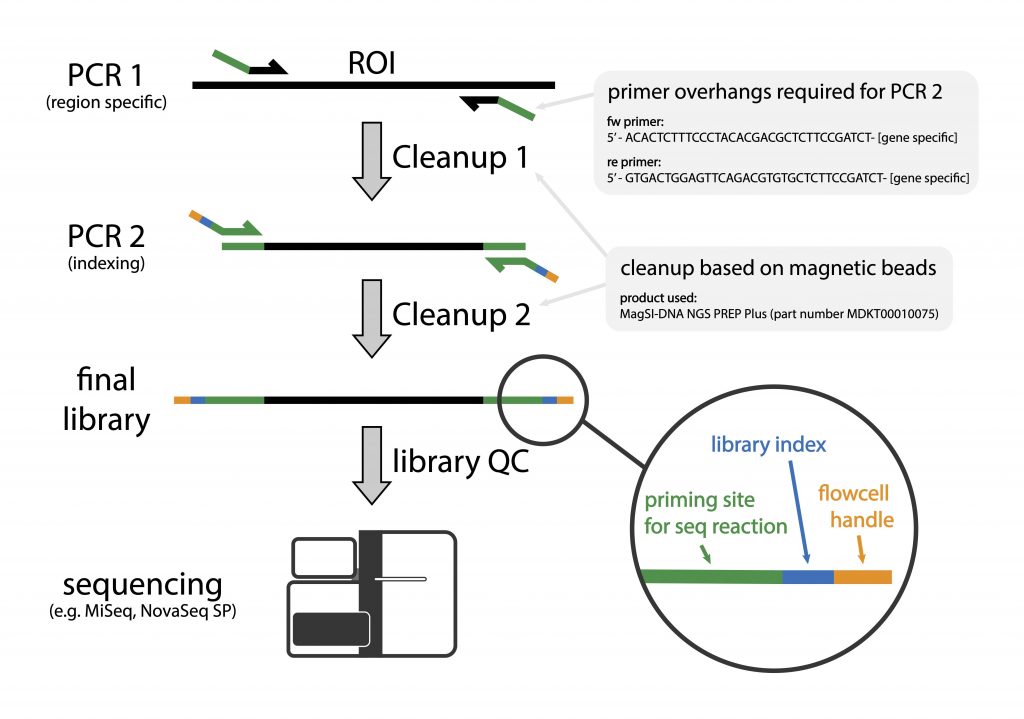
Library preparation is carried out in two PCR-steps. The first PCR involves the use of locus-specific primers (341F/805R) that target the hyper-variable part (V3/V4) of the bacterial 16S gene. This primer pair has 5’-overhang handles that enable the introduction of sample-specific index sequences via a second PCR.
While the protocol offered by NGI has been optimised for a template concentration of 0.25ng/uL of pure bacterial genomic DNA and multiple projects with environmental samples, your samples could contain other sources of DNA than what has been tried. Therefore the template concentration for library preparation for your samples might need to be higher.
Moreover, the presence of secondary metabolites in environmental samples could impact the success of prep since these substances could interfere with the polymerase used for PCR.
It is therefore important to take these factors into account prior to starting a project. Please discuss your project with us prior to placing an order.
Test PCR prior to submitting samples to NGI
Before starting the project we would like you to perform a test PCR (PCR1) on a few of your samples (ideally 10 samples per 96-well plate submitted to NGI).
Please include samples that might be very different from one another for e.g. from slightly different sources and where you would expect variable amounts of bacterial gDNA. PCR1 is an important step because it helps to infer the content of bacterial gDNA in the respective sample, which could serve as a reference for determining the concentration that samples should be normalised to prior to being delivered to NGI.
We will send you a requisite amount of primers based on the number of samples in your project.
The primers will be sent to you once your project has been set up by us or after you have had a meeting with one of our Project Coordinators to discuss your specific project.
Sample requirements
- Sample Type: gDNA
- Sample Amount: All samples submitted to us should have the same concentration in ng/ul i.e. they should be normalised. This is essential since we use a set primer concentration and PCR conditions for all samples in a given plate. The actual concentration to submit needs to be determined by doing a test PCR, see instructions below.
- Sample Volume: 14 μl.
- Sample QC: Sample concentrations MUST be measured fluorometrically for e.g. on a Qubit instrument with the HS DNA kit (Qubit™ 1X dsDNA High Sensitivity (HS) Thermo Fisher Scientific, catalog number Q33230 (100 assays) or Q33231), and be normalised prior to delivery. We will be unable to work with material where concentration was estimated by spectrophotometry. Contact us if you have any questions.
- Maximum number of samples/plate: 94, leave the last 2 wells in the last column (G12 & H12) EMPTY (please do not add water or buffer). We add our controls to these wells.
Conditions for test PCR1
Input DNA concentration: The required template concentration could vary depending on the fraction of bacterial DNA in the samples. It is therefore required that you perform a test PCR with different input concentrations (from e.g. 0.25ng/uL to ca 5ng/uL).
Primers: Aliquots of primers we send you for test PCR1. Questions? Contact us!
Polymerase: Please use KAPA polymerase (KAPA HiFi HotStart ReadyMix PCR Kit; KK2602, Roche) for your test PCR since it is what we will be using at NGI during prep.
Reaction set up:
– 4 μL sample
– 10μL KAPA 2x
– 0.5μL BSA (20mg/uL)
– 4.5μL water
– 2μL Fwd primer and Rev primer (mixed – provided by NGI)
Cycling conditions:
1. Denaturation @ 98C°, 2min
2. Denaturation @ 98 C°, 20 s
3. Annealing @ 54˚C, 20 s
4. Elongation @ 72C°, 15 s
5. GO TO step 2, 28 cycles
6. Final elongation @ 72C°, 2 min
Analysing results of the PCR:
Run products on a 1% agarose gel. There should ideally be one band visible at approx. 500bp. The lowest concentration of input that gives this clear band gives an estimate of what concentration your samples should be normalised to prior to delivery. Thereafter, please measure the concentration of this sample on the Qubit to enable normalisation of all the samples. All samples need to be at the same concentration in the plate submitted to us.
Send us an image of the agarose gel prior to shipping us any samples.
Evaluating sample quality
NGI will process all samples on a given plate with the same conditions. We use 4uL of each sample as an input for PCR1 using almost the same cycling conditions described earlier (we use fewer cycles since an additional PCR will be done to add sample barcodes and Illumina adapters).
This means that if the samples on a given plate have varying concentrations due to poor normalisation or differences in the amount of bacterial gDNA varies among individual samples, some samples may amplify badly, while others might be over amplified, severely affecting the results of the study.
We therefore rely on your reported sample concentrations that should be measured fluorometrically.
We also require you to send us the results of your test PCR1 either after agarose gel electrophoresis or through measurements on a TapeStation/Fragment Analyzer.
Gel photos or electropherograms must be clearly labelled with what template DNA concentration was used.
What we do with your samples
Library preparation
We begin by performing PCR1 that specifically targets the 16S rDNA component of the bacterial gDNA content of your sample, assuming that all samples have been normalised to have a similar content of bacterial gDNA. We subsequently purify the PCR1 product using MagSI beads during which the leftover adapters are removed and the sample is concentrated.
This purified product is used as a template for PCR2, which adds the index sequences with the Illumina flowcell adapters. This is followed by another round of bead-based library purification.
We run a positive and negative control on every plate. The positive control we use is pure bacterial gDNA.
Library QC and sequencing
Libraries will be queued directly for sequencing without notifying the user as long as >70% of the samples pass library QC.
Concentration is measured for all libraries, while the size distribution is checked on a subset to confirm that a product with the expected size has been amplified. Libraries with a concentration of >1ng/uL will be considered “passed”.
The sequencing will be carried out following the setup stated in the agreement.
Expected results
While we have successfully generated and sequenced libraries from many different sample types, as previously stated, success at library preparation is strongly dependent on the concentration of bacterial gDNA in the input material. Furthermore, since different sample types might have issues due to PCR inhibitors in the samples, these can render the samples unsuitable to work with and we cannot guarantee success. Since we include a positive control in all our worksets, we rely on that as a measure to determine the success of library preparation for all samples in a specific workset.
Bioinformatics
Last Updated: 18th January 2024